Breadcrumb
- Home
- Research
Research Interests
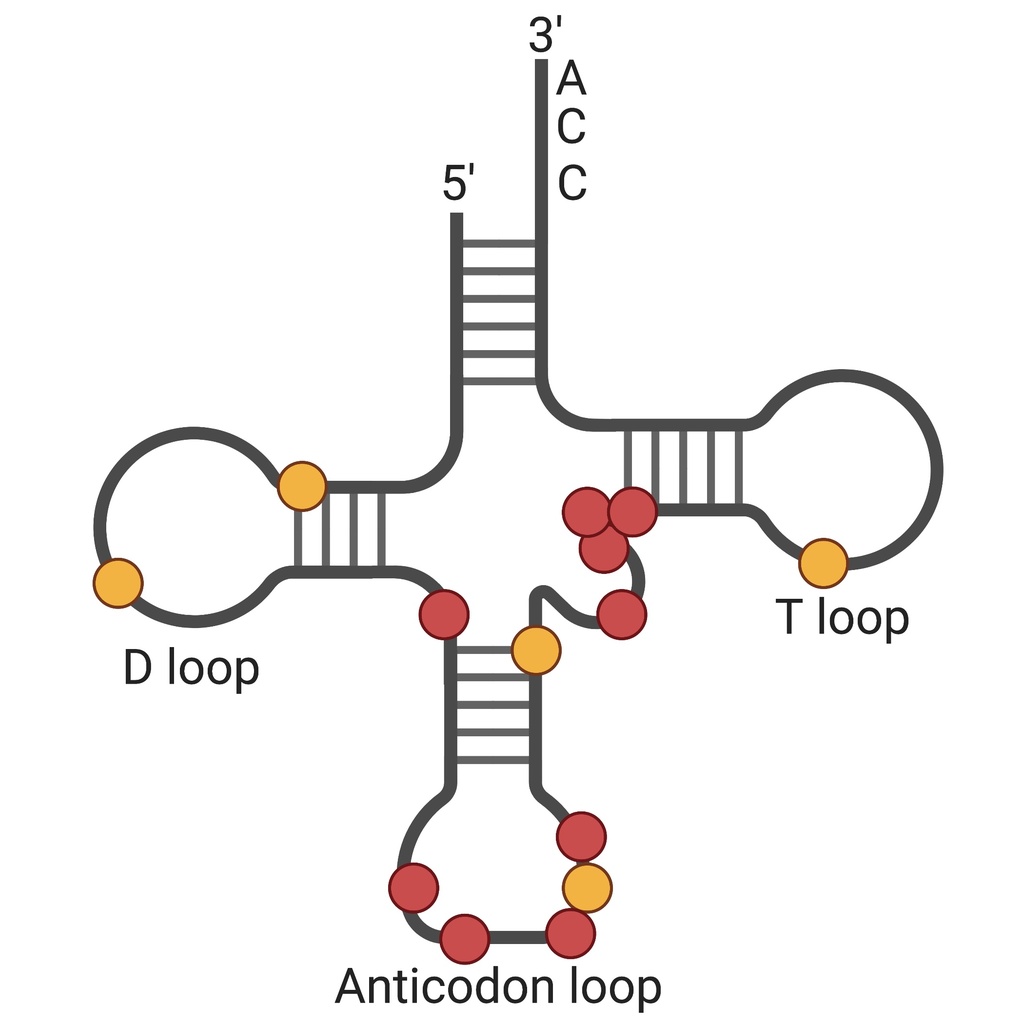
transfer RNA
tRNA sequencing
The discovery of non-coding RNAs, over 60 years ago, began with the identification of the transfer RNA (tRNA), solidifying the central dogma of biology by defining the adaptor for protein translation. However, tRNA expression and regulation has proved exceptionally hard to dissect—a major barrier to our understanding of tRNA biology is the fact that tRNAs are heavily modified, making it difficult to perform accurate high-throughput tRNA sequencing. These technical challenges have resulted in substantial gaps in our knowledge of mammalian tRNA expression, particularly the mechanistic details regarding tRNA processing, modification, and turnover. We are interested in applying new sequencing technologies to improve our ability to analyze tRNA expression in response to viral infection and beyond.
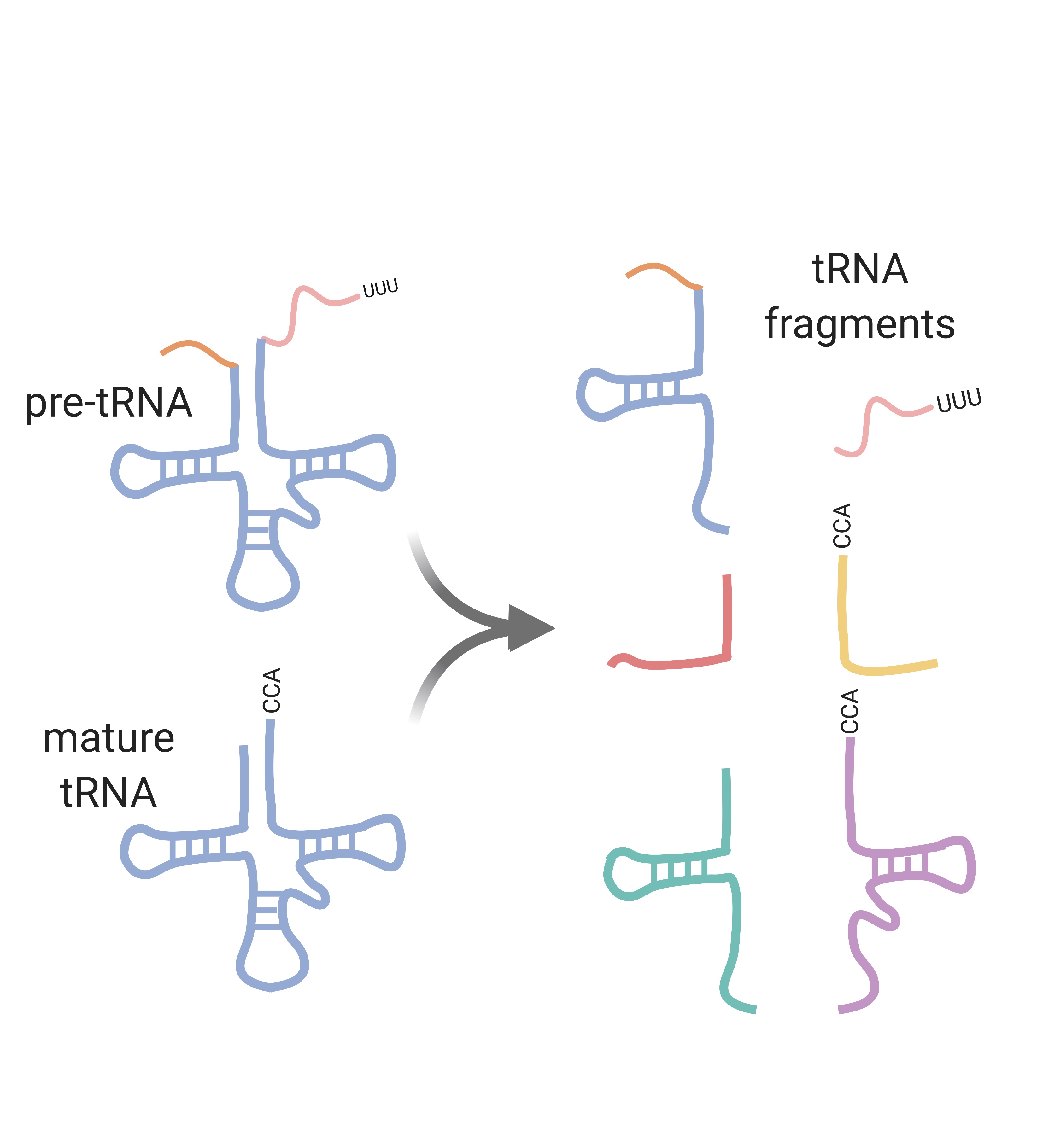
Infection-induced tRNA fragments
tRNA fragments have been implicated in the regulation of gene expression under diverse biological conditions, yet our understanding of this novel class of small RNAs remains limited. Evidence suggests that tRNAs are cleaved into tRNA fragments in response to stress and viral infection, and that tRNA fragments impart functional consequences in cells through diverse translational control mechanisms. We are identifying and functionally characterizing gammaherpesvirus-induced tRNA fragments, ultimately gaining perspective on whether tRNA fragments are weapons waged by the virus or the host.
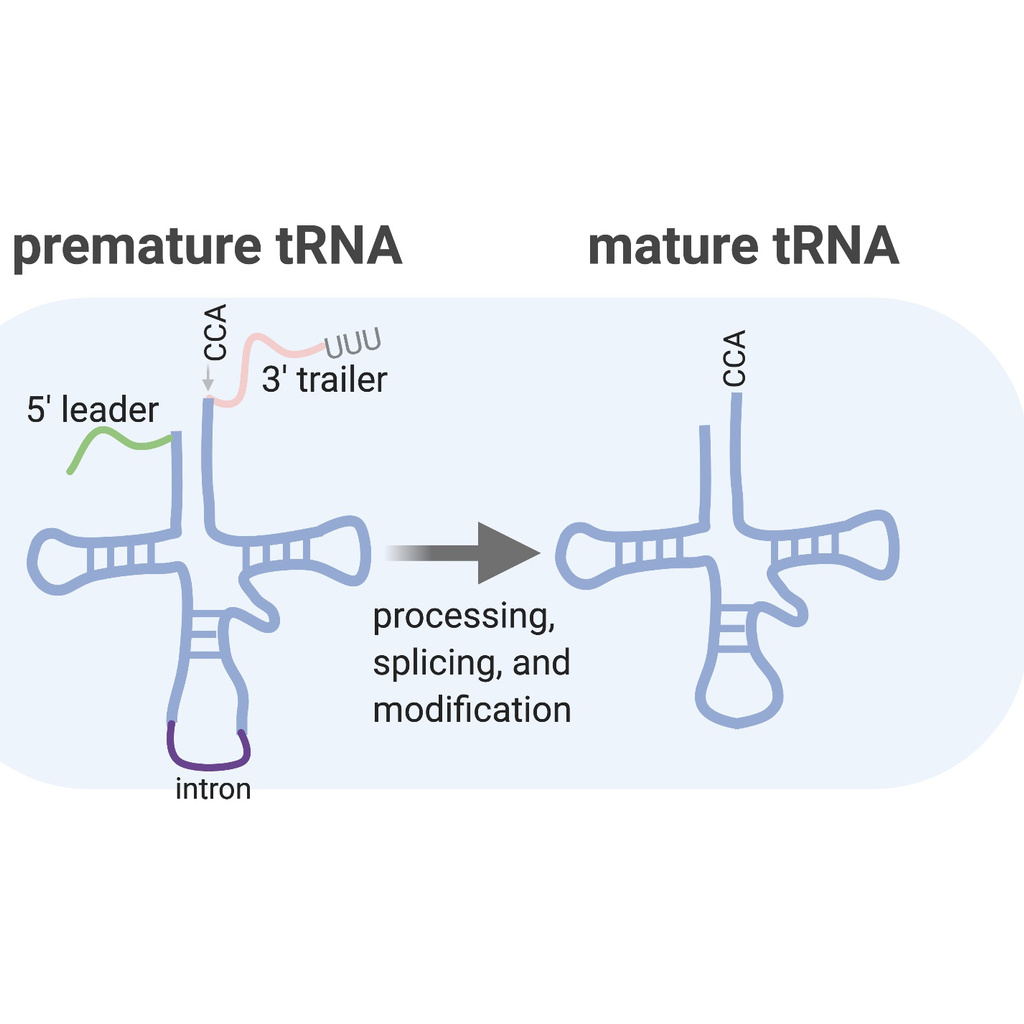
Dissecting tRNA maturation
tRNAs are complex small RNAs—they must undergo many rounds of modification, trimming, and cleavage to produce a functional, mature tRNA molecule. We have found that gammaherpesvirus infection leads to a block in normal tRNA maturation, leading to increased expression of premature tRNAs. We are interested in identifying the host and/or viral factors involved in this maturation block. Our characterization of tRNA-related factors will be highly relevant to our understanding not only of gHV replication, but also of neurological and metabolic disorders associated with defects in normal tRNA biology.